- [课程通道] Gene Interference and Cat Allergen Silencing Research Project
- [课程通道]
- [科学少年] 2025-10-05
- [科学少年] 2025-10-02
- [科学少年] 2025-10-01
CRISPR Is Not Limited to 爱博物 2025-09-18
[Foreword]
This article is a submission from Li Chunman, a student who participated in the IBOWU · JSR Academy 2023 summer course on Zebrafish Embryo CRISPR Gene Editing at the National Marine Gene Bank. The manuscript is excerpted from her final project presentation. For her presentation, she chose to explore a more in-depth research direction related to CRISPR, which is highly commendable. We welcome more submissions from IBOWU students.
CRISPR Is Not Limited to "God's Scalpel"
By Li Chunman (Mandy)
At the end of June this year, I participated in the IBOWU · JSR Academy Zebrafish CRISPR Gene Editing advanced customized research program. During this project, we systematically learned the basic theory and experimental procedures of CRISPR and completed everything from the design and preparation of sgRNA to the final microinjection of zebrafish embryos.
For the final presentation topic, I chose: "Using CRISPR to cause high/low expression of a gene without causing DSB (Double-Strand Breaks)." This was a completely new challenge for me.
In the CRISPR system, the technology used to cause high expression of a gene without causing DSB is called CRISPRa, and a similar technology used for knockdown is called CRISPRi. Because these two technologies do not affect the actual gene sequence, many publications categorize them as epigenetic editing or pseudo-evolutionary editing. Both CRISPRa and CRISPRi fall under CRISPR-dCas9-EE technology, where EE stands for Epigenetic Editing. The 'a' in CRISPRa stands for 'activation,' used for gene knock-up; the 'i' in CRISPRi stands for 'inhibitor' or 'interference,' used for gene knock-down. Let me introduce these two technologies.
Both CRISPRa and CRISPRi systems use the dCas9 protein to "activate" or "block" gene expression at the transcription stage. Both systems require three components: an sgRNA, a dCas9 protein, and an activator or inhibitor. The sgRNA, as in the standard CRISPR system, is used to guide the Cas9/dCas9 protein. The dCas9 protein stands for 'dead Cas9' or 'endonuclease-deficient Cas9,' meaning it is an inactive Cas9 protein. dCas9 is formed by inactivating the HNH and RuvC catalytic domains of a normal Cas9 protein through point mutations. As shown in (Figure 1), inactivating the HNH and RuvC catalytic domains is like removing the two scissors of the Cas9 protein that cut DNA, preventing dCas9 from causing DSBs.
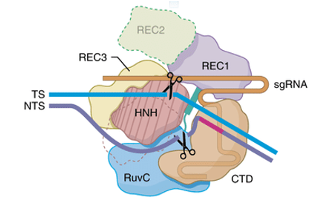
Figure 1: DNA structure (Xing, Z et al., 2019)
CRISPRa technology is quite simple; it involves fusing dCas9 to an Activator. Activator(s)/Transcriptional Activator(s) work by recruiting RNA polymerase II (the machinery for mRNA production) and Transcription Factors (TFs) to the promoter region, causing that segment of DNA to be transcribed into mRNA repeatedly, resulting in high expression of the gene. Typically, Activators contain two regions: one that binds to dCas9 and another responsible for activating transcription. CRISPRa technology also includes several different systems.
The most original and simple system is called VP64 (Figure 2). This system fuses a single VP64 to dCas9; VP64 is the domain from the GCN4 protein responsible for recruiting TFs. The VPR or VP64-p65-Rta system (Figure 3) builds upon the VP64 system by adding two additional TFs, p65 and Rta, to VP64; more transcriptional factors make this system more effective. A relatively popular system is called SAM (Synergistic Activation Mediator) (Figure 4). It adds MS2 to the VP64 base; the RNA片段 within MS2 binds to the sgRNA causing it to fold, and can recruit p65 and HSF1 (two TFs). Compared to VPR, this system is more complex but more efficient. The final system I will introduce is called SunTag (Figure 5). This system adds more VP64 units to form a long chain, making the gene knock-up effect more pronounced.
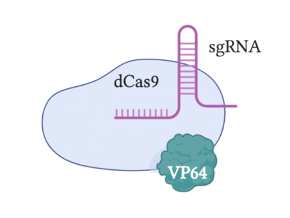
Figure 2: CRISPRa VP64
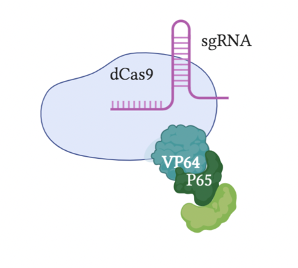
Figure 3: CRISPRa VPR
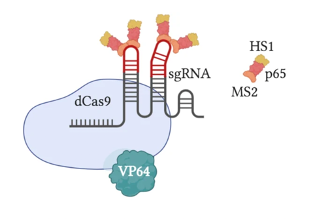
Figure 4: CRISPRa SAM
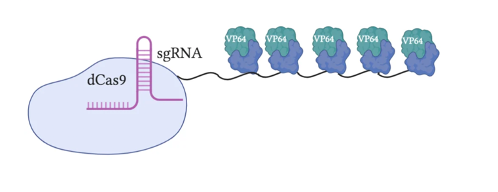
Figure 5: CRISPRa SunTag (Clouse, 2020)
CRISPRi technology is almost the same as CRISPRa, except an inhibitor is fused to dCas9 instead of an activator. The inhibitor/repressor domain fused to dCas9 also comes in various systems. In fact, dCas9 alone can cause gene knockdown by binding to the promoter and preventing RNA polymerase II from accessing the DNA for transcription into mRNA. However, the success rate and efficiency using just dCas9 are relatively low, so most experiments using dCas9 for knockdown add an inhibitor. A well-known CRISPRi system is called KRAB-ZFP. KRAB-ZFP is the abbreviation for Krüppel-associated box - Zinc finger proteins. KRAB is a member of the largest family of transcriptional repressors encoded in the genomes of higher organisms. The KRAB domain is located at the end of the KRAB-ZFP system and communicates with the scaffold protein KAP-1. KRAB has two methods to suppress gene expression: The first uses KAP-1, which can recruit various transcription factors to suppress gene transcription.

The second method is through heterochromatinization; this involves bringing histones closer together to prevent mRNA transcription. The KRAB-ZFP system (Figure 6) includes, besides KRAB and KAP-1, Mi2a which removes acetylation from histone surfaces (acetylation via lysine residues gives histones a negative charge, repelling the negatively charged DNA), SETDB1 which adds methyl groups to histones (methylation can block/inhibit mRNA transcription), DNMT1 which carries information for adding methyl groups, and HP1 which binds methyl groups and pulls histones closer together.
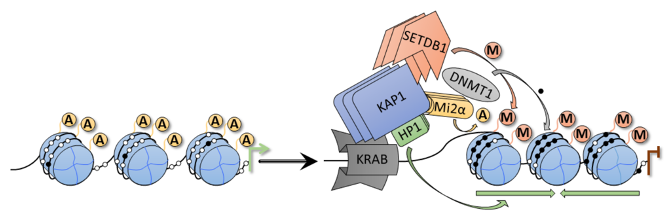
Figure 6: KRAB-ZFP structure
Using CRISPRa and CRISPRi has many advantages; it is a reversible form of gene editing and does not affect the DNA sequence, thus avoiding ethical concerns. It also makes studying gene function more convenient because, like a switch, it allows researchers to arbitrarily regulate gene expression, unlike standard CRISPR which requires complex cutting and pasting. Finally, this type of editing does not affect the p53 protein. p53 is a protein that prevents cancerous mutations; upon detecting a mutation, p53 eliminates the cell to prevent the mutation from being inherited/propagated through cell division; cell elimination leads to unsuccessful gene editing. Therefore, CRISPRa and CRISPRi are more likely to succeed than standard CRISPR gene editing.
References:
Boyle, Patrick, and Charles Després. “Dual-Function Transcription Factors and Their Entourage.” Plant Signaling & Behavior, vol. 5, no. 6, June 2010, pp. 629–634, https://doi.org/10.4161/psb.5.6.11570.
Clouse, Gabrielle . “DCas9-VP64,” CRISPR Activators: A Comparison between DCas9-VP64, SAM, SunTag, VPR, and More!, 2020, blog.addgene.org/crispr-activators-dcas9-vp64-sam-suntag-vpr#:~:text=Guided%20by%20dCas9%2C%20VP64%20recruits,on%20which%20sequence%20is%20targeted.
“CRISPR Activation.” Wikipedia, 28 Mar. 2023, en.wikipedia.org/wiki/CRISPR_activation#:~:text=To%20overcome%20the%20limitation%20of. Accessed 21 July 2023.
“CRISPR-Based Synergistic Activation Mediator (SAM).” Bpsbioscience.com, bpsbioscience.com/crispr-based-synergistic-activation-mediator-sam. Accessed 21 July 2023.
“CRISPRa: A Simpler Way to CRISPR Gene Activation.” Bitesizebio.com, 4 Oct. 2019, bitesizebio.com/44248/crispr-based-activation-crispra-of-genes-a-how-to-guide/. Accessed 21 July 2023.
Lupo, Angelo, et al. “KRAB-Zinc Finger Proteins: A Repressor Family Displaying Multiple Biological Functions.” Current Genomics, vol. 14, no. 4, 1 June 2013, pp. 268–278, https://doi.org/10.2174/13892029113149990002.
Ma, Jun. “Transcriptional Activators and Activation Mechanisms.” Protein & Cell, vol. 2, no. 11, Nov. 2011, pp. 879–888, https://doi.org/10.1007/s13238-011-1101-7.
“Synthego | Full Stack Genome Engineering.” Www.synthego.com, www.synthego.com/guide/crispr-methods/crispri-crispra#:~:text=What%20is%20CRISPRa%3F. Accessed 21 July 2023.
Wang, Chengkun, et al. “DCas9-Based Gene Editing for Cleavage-Free Genomic Knock-in of Long Sequences.” Nature Cell Biology, vol. 24, no. 2, 1 Feb. 2022, pp. 268–278, www.nature.com/articles/s41556-021-00836-1, https://doi.org/10.1038/s41556-021-00836-1. Accessed 8 May 2022.
If you want to learn more and delve deeper into the mysteries of gene editing, welcome to come to the Qingdao National Marine Gene Bank and, like a scientist, personally complete CRISPR experiments!
For more detailed information, please consult an IBOWU course advisor.
Course Advisors:
Teacher Zhang: 13910908618 (WeChat ID same as phone number)
Teacher Zha: 13910312020 (WeChat ID same as phone number)
About IBOWU · JSR Academy
JSR Academy™ is China's first junior science academy initiated by non-governmental forces. Launched by IBOWU, a well-known domestic platform for youth scientific research and academic programs, it aims to provide high-quality scientific practice platforms for Chinese teenagers.
IBOWU has received investments from BGI Group, TAL Education Group, and Poly Group. Leveraging strong scientific and capital backgrounds, it has gathered dozens of multidisciplinary frontline scientists and resources from joint laboratories distributed across dozens of universities and research institutions in multiple countries including China, the US, Denmark, Norway, Singapore, and Australia. It is committed to providing students with high-end STEM advanced academic programs and elite training plans, striving to cultivate the scientific spirit of Chinese youth and reserve talent for China and the global scientific and technological community.



